Gene Expression Analysis with R: Applications in Cancer, Immunology, and Beyond
Register for Gene Expression Analysis using R Masterclass where you will learn:
- Poor Quality or Noisy Data
- Inadequate Sample Size
- Batch Effects and Technical Variability
- Incorrect Normalization of Gene Expression Data
- Choice of Statistical Methods
- Multiple Testing Correction Issues
- Annotation and Interpretation Challenges
- Functional Enrichment Analysis Pitfalls
- Lack of Biological Replicates
- Difficulty in Integrating Multi-Omics Data

Demo Session: MARCH 28-29, 2025; 10:00 AM IST Onwards
To Be Announced Soon
An immersive, hands-on masterclass filled with powerful tools and frameworks to master Differential Gene Expression Analysis using R 🧬. Gain hands-on experience, uncover biological insights, and elevate your data analysis skills. Join us!
Introduction To the course
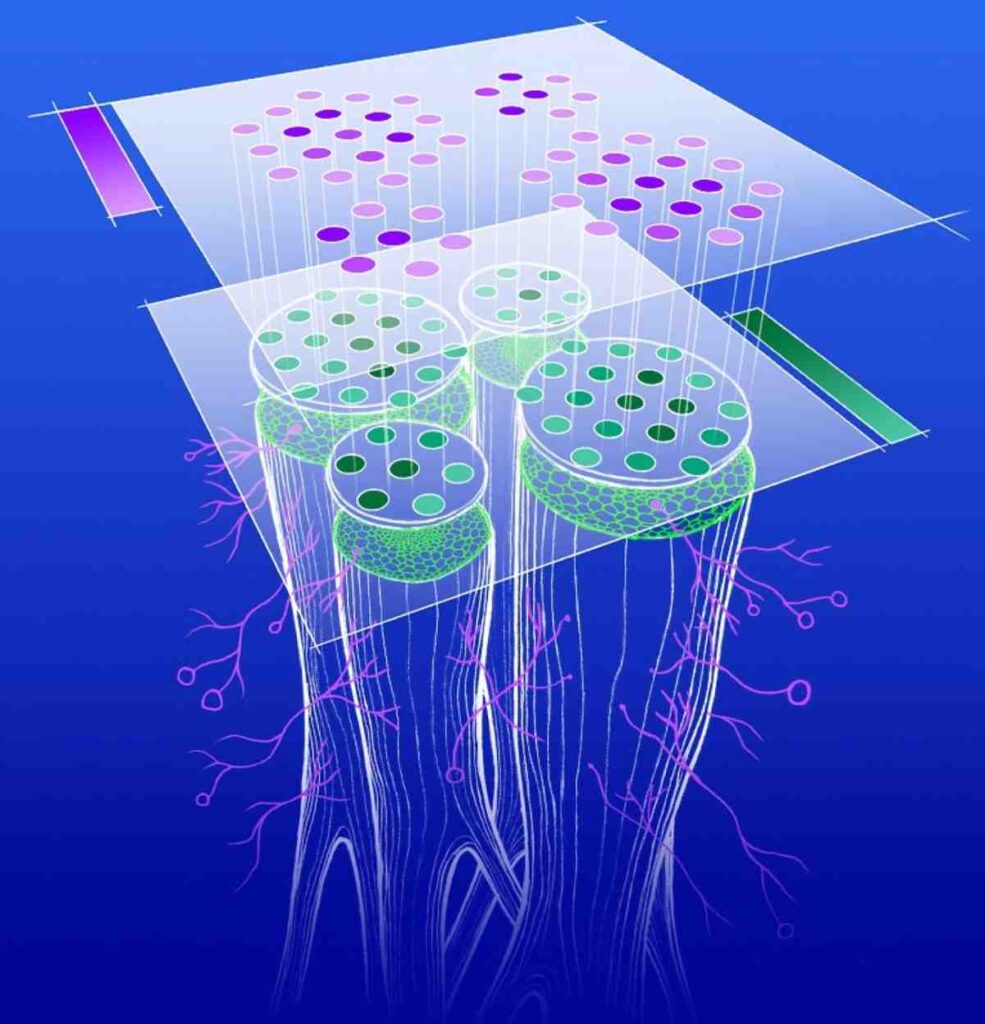
Gene expression analysis is a fundamental technique in bioinformatics, enabling researchers to study how genes are activated or suppressed under different conditions. R, a widely used statistical programming language, provides powerful tools for analyzing gene expression data from RNA sequencing (RNA-Seq) and microarrays. With specialized packages like DESeq2, edgeR, and Limma, researchers can perform essential steps such as data preprocessing, normalization, differential expression analysis, and functional enrichment.
This workshop introduces key concepts, guiding participants through hands-on exercises to identify differentially expressed genes, create heatmaps, volcano plots, and interpret biological insights. Understanding gene expression patterns is crucial for fields like cancer research, drug discovery, and precision medicine. By leveraging R’s extensive bioinformatics libraries, researchers can process large-scale transcriptomic datasets with reproducible workflows.
Whether you’re a beginner or looking to refine your skills, this session provides a strong foundation for advanced gene expression studies in bioinformatics and computational biology.
CONTENTS OF THIS 2 DAY COURSE (TOPICS COVERED)
DAY 1 - Session 1 (10:00 AM - 11:00 AM)
1 Hr 00 Minutes
Introductory Session
Participants will Understand the Core Concepts of the Technology, including different parameters used and Terminologies.
DAY 1 - Session 2 (11:15 AM - 01:15 PM)
2 Hr 00 Minutes
Data Pre-Processing
We will Download and check the Quality of the data and remove bad-quality Reads, including anomalies, noise, duplicate data, Etc.
DAY 1 - Session 3 (02:00 PM - 04:00 PM)
2 Hr 00 Minutes
Mapping with Reference genome
Downloading the reference genome, building the index for mapping, then mapping our genome with the reference genome & analysing the mapping.
DAY 1 - Session 4 (04:15 PM - 06:00 PM)
1 Hr 45 Minutes
Post-Mapping Processing
Once the mapping is completed we will perform the extraction, filtering and restoring process of the mapped reads from the un-mapped reads.
DAY 2 - Session 1 (10:00 AM - 11:00 AM)
1 Hr 00 Minutes
Counts File
We will then use the filtered mappings file for creating the counts file and then running the analysis on it. In order to generate the data for analysis with R.
DAY 2 - Session 2 (11:15 AM - 01:15 PM)
2 Hr 00 Minutes
R for Gene Expression Analysis
We will now use the data generated with the counts file and run the gene expression analysis by normalising the data first & Generate the Results.
DAY 2 - Session 3 (02:00 PM - 04:00 PM)
2 Hr 00 Minutes
Clustering & Pathway enrichment analysis
Now that the results are generated we will now proceed towards performing Clustering Analysis along with GO & KEGG Pathway Analysis.
DAY 2 - Session 4 (04:15 PM - 06:00 PM)
1 Hr 45 Minutes
Visualization & Result Interpretation
We will then generate different graphs like heatmap, violin plot Etc. for result understanding the results and interpreting the outcomes & Concluding the Course.
Demo Session: MARCH 28-29, 2025; 10:00 AM IST Onwards
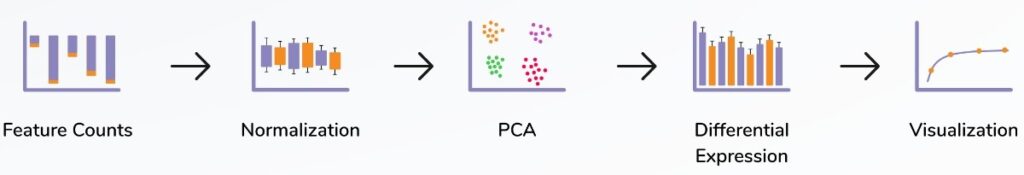
DIFFERENT TYPES OF ANALYSIS
Hi! I’m Jhon Doe. And thanks for taking an interest in reading about me!

Differential Gene Expression (DGE) Analysis
- Identifies genes that are significantly upregulated or downregulated between different conditions (e.g., disease vs. control).
- R Packages:
DESeq2,edgeR,Limma - Example: Finding genes responsible for drug response in cancer samples.
Gene Set Enrichment Analysis (GSEA)
- Determines whether predefined gene sets (e.g., pathways) are significantly enriched in a dataset.
- R Packages:
clusterProfiler,fgsea,GSEABase - Example: Identifying biological pathways altered in a disease state.


Single-Cell RNA-Seq Analysis (scRNA)
- Analyzes gene expression at the single-cell level to study cellular heterogeneity.
- R Packages:
Seurat,SingleCellExperiment,monocle3 - Example: Understanding immune cell diversity in tumor microenvironments.
Demo Session: MARCH 28-29, 2025; 10:00 AM IST Onwards
EXPECTED OUTCOMES OF THE COURSE

Identification of Differentially Expressed Genes (DEGs)
- Detect upregulated and downregulated genes between conditions (e.g., healthy vs. diseased samples).
- Helps in understanding gene regulation and disease mechanism.

Gene Ontology (GO) and Pathway Enrichment Analysis
- Identify biological processes, molecular functions, and pathways associated with DEGs.
- Helps in functional annotation using tools like cluster Profiler.

Visualization of Gene Expression Data Patterns
- Generate heatmaps, volcano plots, and PCA plots to explore expression differences.
- Helps in clustering samples based on expression profiles.

Biomarker Discovery for Disease Diagnosis
- Identify potential gene biomarkers linked to diseases or treatment responses.
- Useful in precision medicine and drug development.

Single-Cell Gene Expression Analysis Insights
- Analyze gene expression at the single-cell level to study cellular heterogeneity.
- Helps in identifying cell types and subpopulations in tissues.

Reproducible and Scalable Transcriptomic Data Analysis
- Standardized pipelines in R ensure reproducibility and scalability for large datasets.
- Enables meta-analysis and integration with multi-omics data.
Demo Session: MARCH 28-29, 2025; 10:00 AM IST Onwards
TAKE AWAY FROM THE COURSE
Preparatory Pre-Recorded Course
To guide you through some of the concepts required to be known by all the participants before joining the course so that everyone is on the same page when the training starts.

Participation Certificate
Industry Recognised Course completion certificate to showcase that you successfully completed the course and submitted all the assignments as well.

Session Recording
Once the training is concluded you will get access to our LMS portal where you can access all the recording of the process we performed during the session for use later.
Demo Session: MARCH 28-29, 2025; 10:00 AM IST Onwards
Why Should You Learn from Us?
We’ve experienced the most common challenges, hiccups and missing pieces that most people struggle with when it comes to starting a career in Genome Analysis.
After training more than 5000 participants from different backgrounds we have identified common problems and misconceptions around this hence, we’ve condensed my 8+ years of experience into a robust 12-week training program with a strong community to support you through the entire process.
With main being on troubleshooting and not just learning how to use a tool.

FREQUENTLY ASKED QUESTIONS [FAQ]
I am New to Bioinformatics, can i join the course?
Absolutely. The program is designed to help you understand from a fundamental level and build it up to advanced concepts.
Do I need to know coding?
Definitely not. As, we will be learning to code as a part of this course.
What else will I be learning in this course?
Apart from Genome Analysis, you will also be learning Ubuntu, Software tools Installation, R Packages Installation, Problem Solving, and managing deadlines – all of which will be very important for a successful career in Genomics.
Apart from this, our career coach will be training you in creating effective resumes, updating your LinkedIn profile to attract opportunities, and modern job-hunting strategies that can help you land a high-paying Bioinformatics job.
I know a few things included in the course, can i skip them?
No, in order to be eligible for getting the certificate you need to attend all the sessions.
Will I get a certificate?
Yes, you will receive an industry-recognised course completion certificate that you can proudly show to your friends and family. But you will need to attend all the sessions to receive the certificate.
Can I pay the registration fee in cash at the venue on day1?
No, All the registrations are required to be done in advance through online payment as we need to prepare your certificates and share the pre-workshop material with you. Hence, we do not allow SPOT Registrations and do not accept cash payments as well.
Demo Session: MARCH 28-29, 2025; 10:00 AM IST Onwards
<iframe src="https://docs.google.com/forms/d/e/1FAIpQLSemaTxm9K9TPO0NaX1uirPhgKckZzEp1C7MYcMUbVDXpBugQA/viewform?embedded=true" width="640" height="2474" frameborder="0" marginheight="0" marginwidth="0">Loading…</iframe>