Decode the Genome Analysis: Hands-On Masterclass in WGS & GWAS with GATK
Register for Whole Genome Sequecing with GATK & GWAS Masterclass where you will learn:
- Handling Large-Scale Data
- Data Preprocessing Challenges
- Computational Resource Requirements
- Pipeline Optimization & Parameter Tuning
- Variant Calling & False Positives
- Population Stratification Issues
- Multiple Testing & False Discoveries
- Lack of Functional Interpretation
- Integration with Other Omics Data
- Reproducibility & Standardization

Demo Session: APRIL 25-26, 2025; 10:00 AM IST Onwards
To Be Announced Soon
A power-packed masterclass filled with cutting-edge tools and frameworks to master Whole Genome Sequencing with GATK & GWAS 🧬. Gain hands-on experience, analyze real data, and unlock genomic insights. Join us!
Introduction To the course

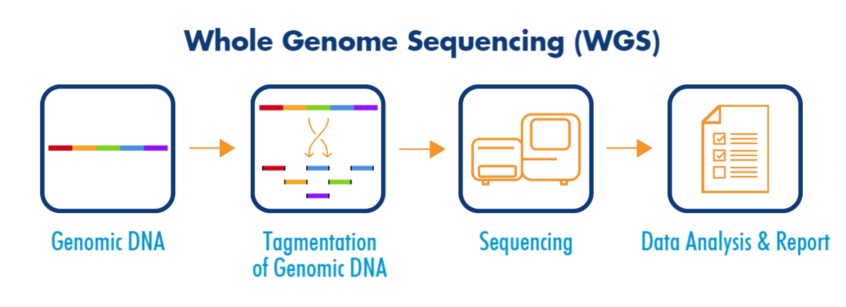
Whole Genome Sequencing (WGS) is a powerful approach for studying genetic variations across the entire genome, providing insights into mutations, structural variants, and disease associations. GATK (Genome Analysis Toolkit) is a widely used tool for processing WGS data, offering pipelines for alignment, quality control, variant calling, and filtering. Genome-Wide Association Studies (GWAS) enable researchers to identify genetic variants linked to specific traits or diseases by analyzing large-scale genomic datasets.
Combining GATK and GWAS allows for high-precision variant detection and statistical association analysis, helping uncover the genetic basis of complex diseases, drug responses, and population genetics.
However, challenges like data preprocessing, computational requirements, and population stratification must be carefully managed to ensure accurate results. This approach is essential for researchers in genomics, precision medicine, and evolutionary studies, enabling them to conduct reproducible and high-quality genomic research for medical and scientific advancements.
CONTENTS OF THIS 2 DAY COURSE (TOPICS COVERED)
DAY 1 - Session 1 (10:00 AM - 11:00 AM)
1 Hr 00 Minutes
Introductory Session
Participants will Understand the Core Concepts of the Technology, including different parameters used and Terminologies.
DAY 1 - Session 2 (11:15 AM - 01:15 PM)
2 Hr 00 Minutes
Data Pre-Processing
We will Download and check the Quality of the data and remove bad-quality Reads, including anomalies, noise, duplicate data, Etc.
DAY 1 - Session 3 (02:00 PM - 04:00 PM)
2 Hr 00 Minutes
Mapping with Reference genome
Downloading the reference genome, building the index for mapping, then mapping our genome with the reference genome & analysing the mapping.
DAY 1 - Session 4 (04:15 PM - 06:00 PM)
1 Hr 45 Minutes
Post-Mapping Processing
Once the mapping is completed we will perform the extraction, filtering and restoring process of the mapped reads from the un-mapped reads.
DAY 2 - Session 1 (10:00 AM - 11:00 AM)
1 Hr 00 Minutes
Variant Calling & Filtering
Variant Calling & Filtering identifies genetic variations like SNPs and InDels from sequencing data & filtering ensures high-quality, reliable variants for studies.
DAY 2 - Session 2 (11:15 AM - 01:15 PM)
2 Hr 00 Minutes
Variant Annotation
Variant Annotation analyzes genetic variants to determine their biological impactaiding in disease research, functional genomics, and precision medicine.
DAY 2 - Session 3 (02:00 PM - 04:00 PM)
2 Hr 00 Minutes
Normality Testing, SNIP & Amputation
Normality tests assess distribution, SNP analysis identifies variants, and imputation predicts missing genotypes, enhancing statistical power in GWAS.
DAY 2 - Session 4 (04:15 PM - 06:00 PM)
1 Hr 45 Minutes
Population Structure & correlation
Population Structure & Correlation analyze genetic diversity and relationships within populations, correcting for stratification in GWAS.
Demo Session: APRIL 25-26, 2025; 10:00 AM IST Onwards
DIFFERENT TYPES OF ANALYSIS
Hi! I’m Jhon Doe. And thanks for taking an interest in reading about me!

Variant Calling & Annotation using GATK
- Identifies Single Nucleotide Variants (SNVs), Insertions/Deletions (InDels), and Structural Variants across the genome.
- Uses GATK tools like HaplotypeCaller, Mutect2 (for somatic mutations), and GenotypeGVCFs to detect genetic variations.
- Annotation tools like
ANNOVARandVEPhelp interpret the functional impact of detected variants. - Example: Identifying mutations associated with genetic disorders.
GWAS for Trait & Disease Associations
- Identifies statistically significant genetic variants associated with specific traits or diseases.
- Corrects for population stratification and multiple testing to avoid false discoveries.
- Tools: Uses
PLINK,GCTA, andSAIGEto analyze large-scale genomic datasets. - Example: Discovering SNPs linked to diabetes or cancer risk


Population Genomics & Evolutionary Analysis
- Studies genetic diversity, ancestry, and evolutionary history across populations.
- Helps in reconstructing population migration patterns and identifying adaptive genetic changes.
- Tools: like
ADMIXTURE,PCA, andFSTto analyze genetic differentiation. - Example: Understanding the genetic basis of human adaptation to high altitudes.
Demo Session: APRIL 25-26, 2025; 10:00 AM IST Onwards
EXPECTED OUTCOMES OF THE COURSE

Identification of Genetic Variants
- Detect Single Nucleotide Variants (SNVs), Insertions/Deletions (InDels), and Structural Variants using GATK.
- Helps in understanding genetic diversity and disease-associated mutations.

Association of Genetic Variants with Traits
- GWAS identifies SNPs linked to complex diseases (e.g., diabetes, cancer, neurological disorders).
- Helps in discovering genetic risk factors and potential drug targets.

Functional Annotation of Variants
- Predicts the biological impact of variants using annotation tools like ANNOVAR, VEP, or SnpEff.
- Helps in prioritizing variants for further research.

Population Structure & Genetic Ancestry
- Studies genetic diversity, ancestry, and evolutionary patterns using GWAS and WGS data.
- Helps in understanding human migration and adaptation.

Identification of CNVs & Structural Variant
- Detects large-scale deletions, duplications, and translocations using GATK’s gCNV pipeline.
- Important for studying cancer genomics and rare genetic disorders.

Development of Personalized Medicines
- Enables precision medicine by identifying genetic markers for drug response and disease susceptibility.
- Helps in tailoring treatments based on an individual's genetic profile.
Demo Session: APRIL 25-26, 2025; 10:00 AM IST Onwards
TAKE AWAY FROM THE COURSE
Preparatory Pre-Recorded Course
To guide you through some of the concepts required to be known by all the participants before joining the course so that everyone is on the same page when the training starts.

Participation Certificate
Industry Recognised Course completion certificate to showcase that you successfully completed the course and submitted all the assignments as well.

SESSION RECORDING
Once the training is concluded you will get access to our LMS portal where you can access all the recording of the process we performed during the session for use later.
Demo Session: APRIL 25-26, 2025; 10:00 AM IST Onwards
Why Should You Learn from Us?
We’ve experienced the most common challenges, hiccups and missing pieces that most people struggle with when it comes to starting a career in Genome Analysis.
After training more than 5000 participants from different backgrounds we have identified common problems and misconceptions around this hence, we’ve condensed my 8+ years of experience into a robust 12-week training program with a strong community to support you through the entire process.
With main being on troubleshooting and not just learning how to use a tool.

FREQUENTLY ASKED QUESTIONS [FAQ]
I am New to Bioinformatics, can i join the course?
Absolutely. The program is designed to help you understand from a fundamental level and build it up to advanced concepts.
Do I need to know coding?
Definitely not. As, we will be learning to code as a part of this course.
What else will I be learning in this course?
Apart from Genome Analysis, you will also be learning Ubuntu, Software tools Installation, R Packages Installation, Problem Solving, and managing deadlines – all of which will be very important for a successful career in Genomics.
Apart from this, our career coach will be training you in creating effective resumes, updating your LinkedIn profile to attract opportunities, and modern job-hunting strategies that can help you land a high-paying Bioinformatics job.
I know a few things included in the course, can i skip them?
No, in order to be eligible for getting the certificate you need to attend all the sessions.
Will I get a certificate?
Yes, you will receive an industry-recognised course completion certificate that you can proudly show to your friends and family. But you will need to attend all the sessions to receive the certificate.
Can I pay the registration fee in cash at the venue on day1?
No, All the registrations are required to be done in advance through online payment as we need to prepare your certificates and share the pre-workshop material with you. Hence, we do not allow SPOT Registrations and do not accept cash payments as well.