"High-Throughput Genome Mapping, De Novo Assembly, Metagenomic Profiling, and Accurate Detection of Genetic Variants for Advanced Research in Genomics, Microbiology, and Precision Medicine."
Shotgun Sequencing is a versatile, high-throughput technique for sequencing entire genomes or complex samples without the need for prior knowledge of specific genes. This method involves randomly fragmenting DNA into smaller pieces, which are then sequenced in parallel using next-generation sequencing (NGS) technologies. The resulting sequence data is assembled into a complete genome or a metagenomic profile using advanced bioinformatics tools. Shotgun sequencing is ideal for applications like de novo genome assembly, metagenomics, microbial community profiling, and identifying genetic variations in complex samples. It offers comprehensive, unbiased insights into genomic structures, mutations, and microbial diversity, empowering research in genomics, microbiology, and precision medicine.
2 Types of Shotgun Sequencing Services
De Novo Genome Assembly
This service is used to assemble genomes from scratch without a reference, making it ideal for sequencing new or previously uncharacterized organisms. It helps researchers uncover novel genetic information, complete genomes, and understand species-specific characteristics. De novo assembly is particularly useful in species discovery, evolution studies, and agriculture.
Metagenomic Shotgun
Focused on analyzing complex microbial communities, metagenomic shotgun sequencing is used to sequence DNA from environmental, clinical, or ecological samples. This method provides insights into microbial diversity, composition, and functional potential without the need to culture individual organisms, making it ideal for microbiome studies, pathogen detection.
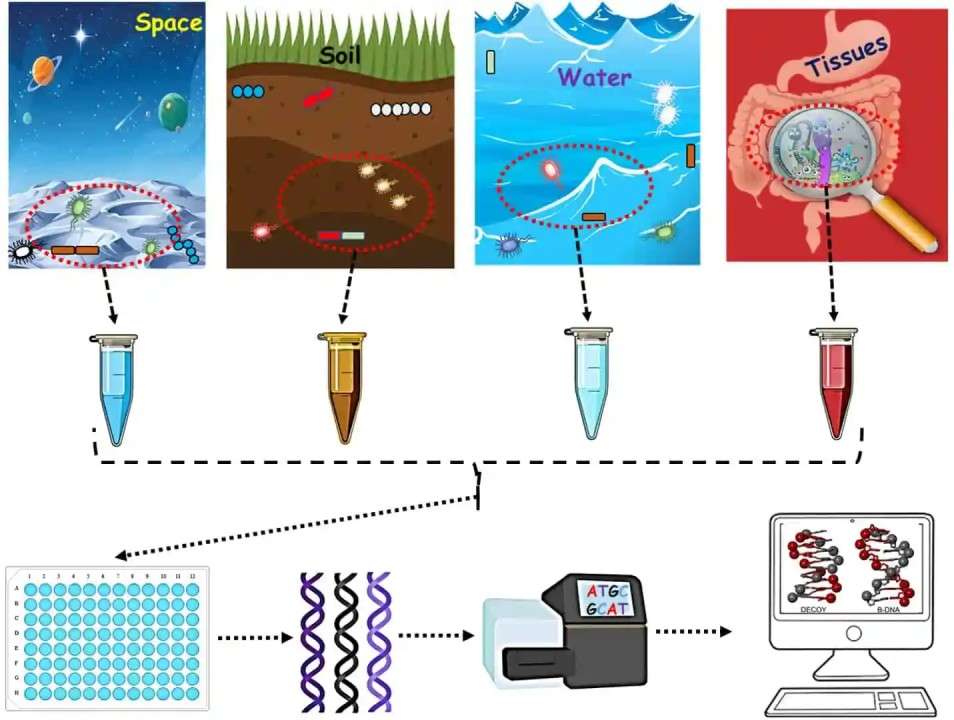
Shotgun sequencing is a powerful, high-throughput method used to analyze entire genomes or complex samples. The process begins with sample collection, which can include environmental samples, tissues, or microbial communities. DNA is extracted from the sample and fragmented into smaller, random pieces, each of which is then sequenced in parallel using next-generation sequencing (NGS) technologies.
Once sequencing is completed, the raw data is processed through advanced bioinformatics pipelines to align and assemble the short reads into a coherent genome or metagenomic profile. This data can then be used for de novo genome assembly to map an organism’s complete genome or to conduct metagenomic profiling for identifying microbial communities.
The final step involves result interpretation, where researchers analyze the assembled data to identify genetic variations, functional genes, and microbial diversity. Shotgun sequencing is essential in areas like genomics, microbiology, disease research, and environmental studies, offering deep insights into complex biological systems.